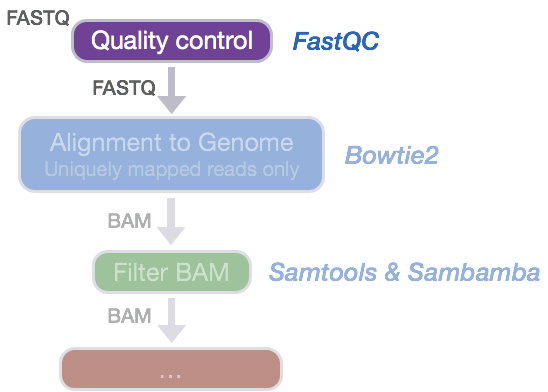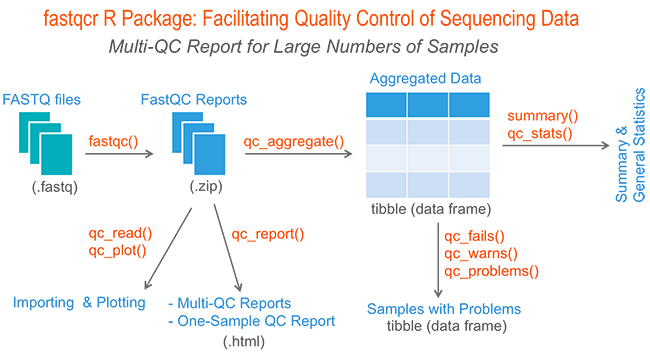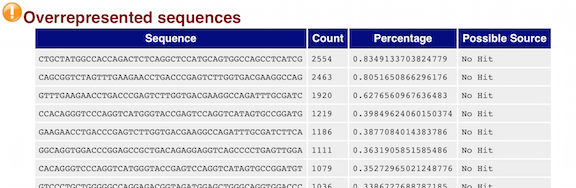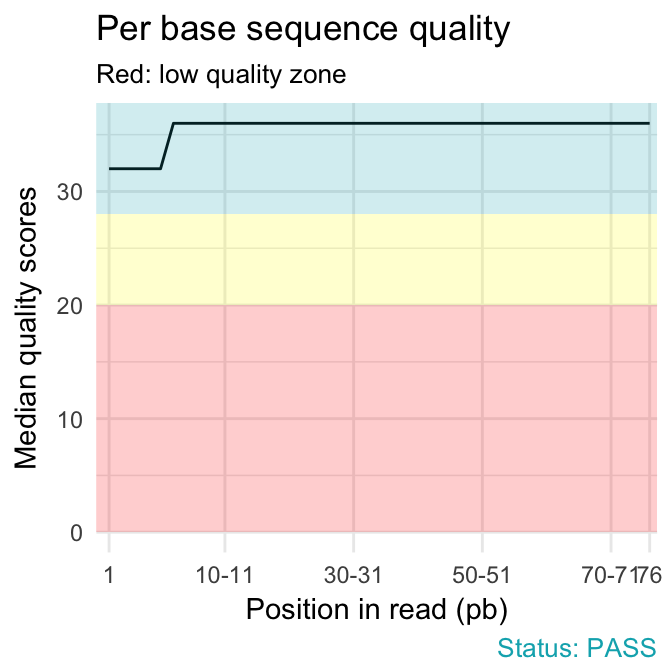
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples - Easy Guides - Wiki - STHDA
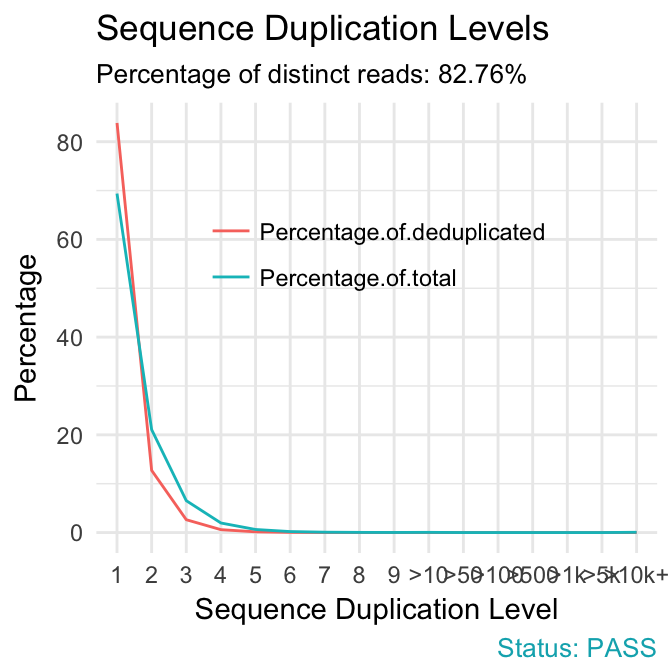
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples - Easy Guides - Wiki - STHDA
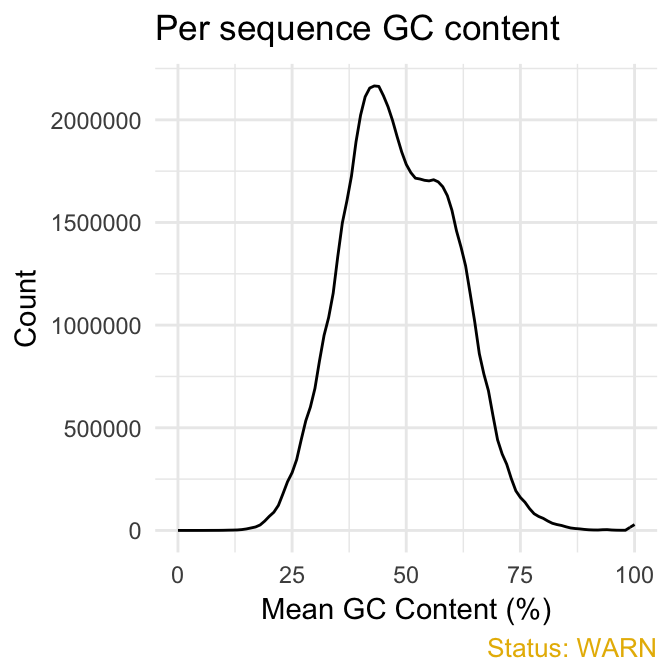
fastqcr: An R Package Facilitating Quality Controls of Sequencing Data for Large Numbers of Samples - Easy Guides - Wiki - STHDA